Section 1 outlines the ingest and clustering processes as parts of SC2 Training Grounds' computer model.
- Load the replay files generated by StarCraft II.
- Call upon the ingest sub-package to inventory the replays and store their relevant indicators into various collections within a document-based database using a local MongoDB client.
- Use the profiler sub-package to build the player profiles from the replay data collections.
- Process the player profiles by game race to build each race classification clusters using the cluster sub-package.
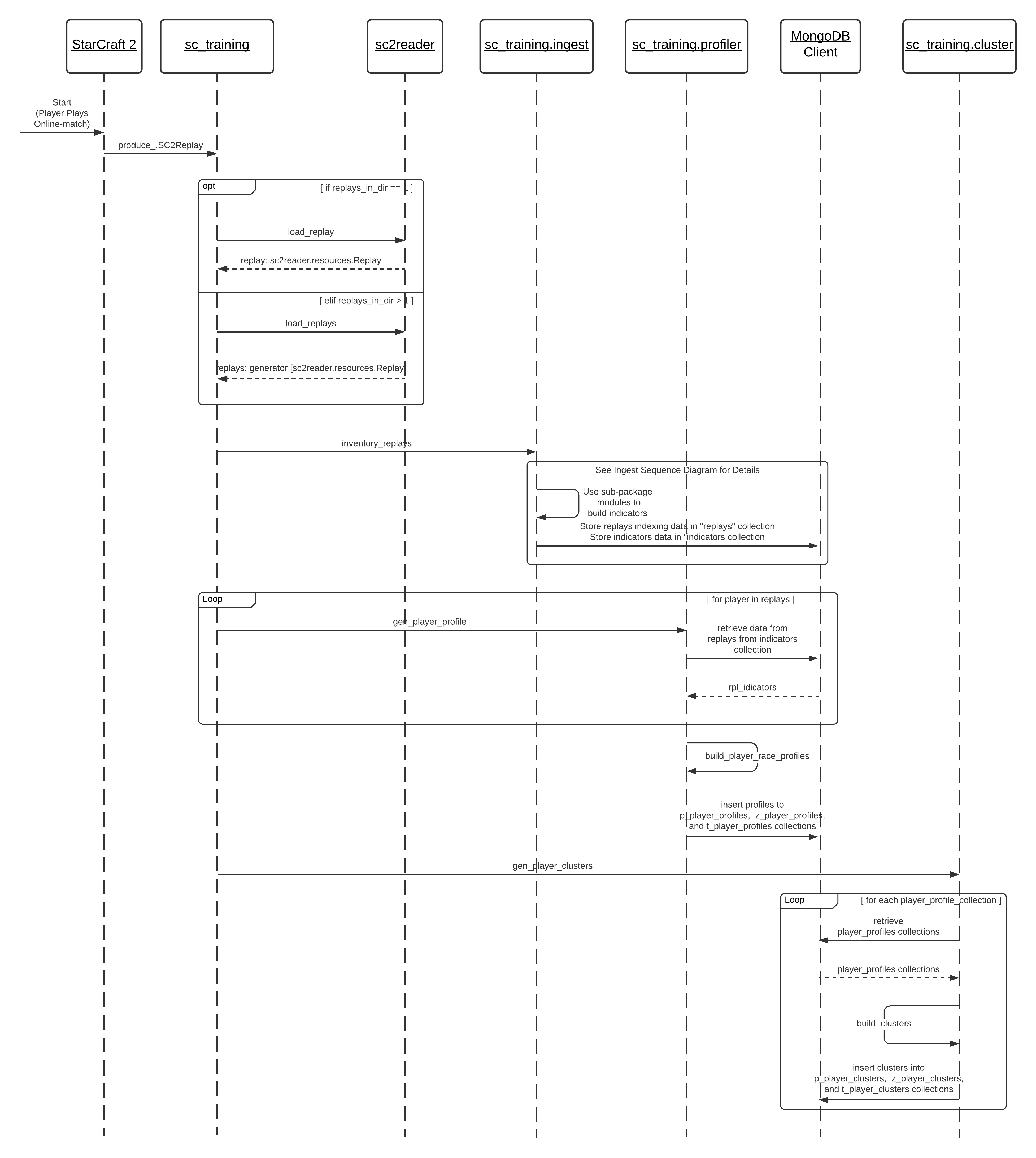
Based on this proposed flow of orders, I first need to implement the ingest process for StarCraft II replay files. This ingestion process is essential to structure the input for the clustering process. Furthermore, it also determines the input of the application's classification modelling process, as shown in SC2 Training Grounds activity diagram (see Section 1's introduction).
In the sequence diagram, I show that the ingestion process is, on a top-level, composed of two main functions, inventory_replays and get_replay_indicators. inventory_replays registers the replays so that a user can use their basic meta-data to reference specific replays and identify their players. Meanwhile, get_replay_indicators compiles all performance indicators of each player in each replay. Thus, the system can loop through various replays and extract and store the performance indicators for each player in each game.
Having understood how the functions are supposed to interact, I implement them in the following modules. The most important outcome of this implementation is the structure of the players' performance indicators and how this data is indexed. In other words, this definition determines the information vectors that will become the inputs for the clustering and classification modelling process afterwards.
The following sequence diagram shows the interaction between these modules in the ingest process.
